June 4, 2019 From rOpenSci (https://deploy-preview-121--ropensci.netlify.app/blog/2019/06/04/rromeo/). Except where otherwise noted, content on this site is licensed under the CC-BY license.
We’ve been following rOpenSci’s work for a long time, and we use several packages on a daily basis for our scientific projects, especially taxize to clean species names, rredlist to extract species IUCN statuses or [treeio](many probs with this post) to work with phylogenetic trees. rOpensci is a perfect incarnation of a vibrant and diverse community where people learn and develop new ideas, especially regarding scientific packages. We’ve also noticed how much the thorough review process improves the quality of the packages that join the rOpenSci ecosystem. And while we were admiring the dynamics of rOpenSci community, we started to wonder how we could contribute to this ecosystem. And this is how we started our quest to find a project that could fit rOpenSci goals while at the same time teach us new skills.
Open Access is the idea that scientific articles should be available to everyone to favour scientific dissemination, as well as public information. Open Access is rising in the scientific community with more and more public funding agencies requiring funded projects to make their article open access. But several very different models exist under this umbrella term of “Open Access”. One of them is the so-called “green open access”, where the articles are made publicly available by their authors via their deposition in institutional or public repositories, such as bioRxiv or HAL. Scientific journals have different policies regarding green open access: some let you archive various versions of the manuscript right after acceptance, while others ask you to wait an embargo period or forbid entirely the archival of the manuscript. To support green open access, scientists can elect to publish their work in journals that authorize manuscript archival. And this is where SHERPA/RoMEO comes in handy: it offers a publicly available database of open access policies of scientific journals and lists the conditions under which manuscript archival might be allowed.
The SHERPA/RoMEO database is available through an Application Programming Interface (API), which meant that we could build an R client to programmatically access this data. This would allow researchers to more easily select journals based on their manuscript archival policies. An R client would also be a precious tool for bibliometricians who want to get the general picture of open access practices in a particular subfield.
Although we had no prior experience working with web data or scraping APIs for data, we knew several tools existed to interact with APIs in R such as httr or rOpenSci’s crul. And as we started the development of rromeo, we also knew we could use the many examples of R client for APIs available on the CRAN Task View on Web Technologies as models.
 SHERPA/RoMEO API
SHERPA/RoMEO API
SHERPA/RoMEO has been available through a web interface since at least February 2004 according to the Internet Archive and their API was released in December 2006 (from the Internet Archive https://web.archive.org/web/20061211125806/http://www.sherpa.ac.uk:80/romeo.php), making their data available to anyone since then.
There are three versions of the manuscript considered in SHERPA/RoMEO:
- pre-print, which is the manuscript version before peer review;
- post-print, which is the manuscript as accepted after peer review but not yet typeset as an article in the journal;
- and typeset manuscript/publisher’s manuscript, which is the manuscript with the text after peer-review, fully typeset, as it appears in the journal.
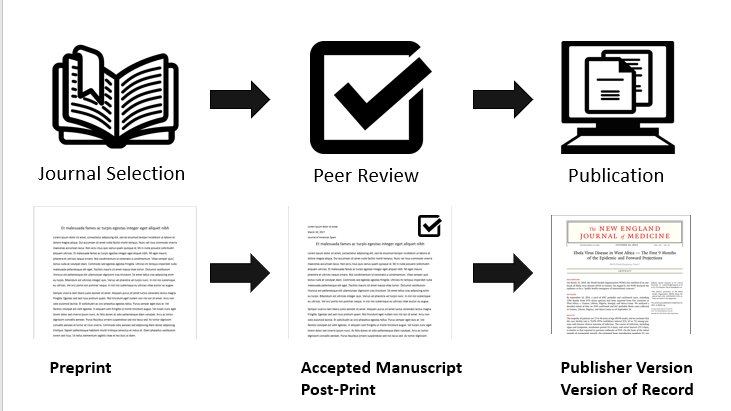
figure by Ryan Regier, with Book icon from Benny Forsberg, CC-BY 3.0
Some journals accept the archival only of the pre-print, while others accept both pre-print and post-print, or even accept the archival of all three versions! SHERPA/RoMEO’s API lets you know what is the policy of a journal using its name, or its ISSN, and whether restrictions apply such as embargo periods before publicly archiving different manuscript versions.
But even though the database is still updated, it seems the development of the API stopped in 2013, which means it’s lacking some functionalities and it does not always follow modern web standards. Because of this, we could not always readily use R packages but we often needed to perform small adjustments first. For example, it did not always use valid XML and the character encoding was not declared in the HTTP headers, but in the body of the document. Furthermore, the SHERPA/RoMEO is not RESTful and thus the queries were a little more complex to design. Fortunately, the developers had written a full documentation of all different types of query we could run.
 rromeo: an R API client
rromeo: an R API client
rromeo lets you access basic information regarding the journal policies in R.
You can get the policy of a specific journal with its title using the function rr_journal_name():
rromeo::rr_journal_name("Methods in Ecology and Evolution")
title issn romeocolour preprint
1 Methods in Ecology and Evolution 2041-210X yellow can
postprint pdf pre_embargo post_embargo pdf_embargo
1 restricted cannot <NA> 12 months <NA>
In this example, we see that ‘Methods in Ecology and Evolution’ allows the archival of pre-print manuscripts, post-print manuscripts (but with restrictions) but it does not allow the archival of the typeset manuscript (pdf column). A 12 months embargo is required before the public archival of post-print manuscripts (post_embargo column).
You can also fetch the policy of several journals at once by matching the beginning of the title via the argument qtype = "starts":
rromeo::rr_journal_name("Bird", qtype = "starts")
4 journals match your query terms.
Recursively fetching data from each journal. This may take some time...
title issn romeocolour preprint postprint
1 Bird Behavior 0156-1383 blue cannot cannot
2 Bird Conservation International 0959-2709 green can can
3 Bird Populations 1074-1755 <NA> <NA> <NA>
4 Bird Study 0006-3657 green can can
pdf pre_embargo post_embargo pdf_embargo
1 can <NA> <NA> <NA>
2 cannot <NA> <NA> <NA>
3 <NA> <NA> <NA> <NA>
4 cannot <NA> <NA> <NA>
Finally, rromeo can also retrieve publisher’s information:
rromeo::rr_publisher_name("Oxford University Press")
romeoid publisher alias romeocolour preprint postprint
1 55 Oxford University Press OUP yellow can restricted
pdf
1 unclear
which gives you the general policy of the ‘Oxford University Press’ regarding manuscript archival. Some restrictions may apply depending on the precise branch of the publisher you plan to publish with, so you should always double check before archiving your manuscripts.
You can get a full overview of rromeo capabilities by reading the introductory vignette.
 Developing an API package: first steps
Developing an API package: first steps
We had no prior experience working with web technologies within R, and this ended up begin a rich and fruitful experience for us. Our first step was to choose the R package to perform the web requests. We picked httr over rOpenSci’s crul package for its even higher-level of use as httr hides even more details on how it handles the query compared to crul. We made sure to follow the best practices described in its “Building API Packages” vignette. We then naturally turned to the xml2 package to parse the resulting XML file, as recommended in rOpenSci package development book.
We wanted to follow best development practices such as having unit tests to check the behavior of functions in the package. But we didn’t intially know how to run unit tests that required an internet connection. We discovered that the answer is mocking: storing locally fake HTTP responses that mimic the API and use them to test the functions in our package.
Creating these fake responses is not an easy task but fortunately, rOpensci’s vcr package is exactly suited for this task. vcr records requests and replays them during the tests (learn more about it in the technote about vcr). We used it in all our tests as well as for caching the examples shown in the README file. The companion book on HTTP testing helped us dive into the different options suitable to record our requests.
While working on rromeo, we realized that the level of details offered to the user was a delicate balance: on one side, we ran the risk of not being flexible enough to be useful and on the other side, the complexity of the package would just become untractable. We were greatly inspired by the many packages developed by Scott Chamberlain that sometimes offer two different interfaces for different kind of users (see rredlist for example). This strategy may be useful to offer different kind of granularity of information on details depending on the future use of the API information.
A similar issue lay with the license of the SHERPA/RoMEO data: the API returns a license notice with each request but we didn’t want to flood the user with too much information. We opted to include this information in the CITATION file of the package accessible with the command citation("rromeo"). We since then noticed other strategies in other packages, such as having a DATA_USE file in the root of the source repository and we are still thinking about what the best choice is here.
 Developing an API package: gotchas and lessons learned
Developing an API package: gotchas and lessons learned
We were eager to learn but we made some mistakes in the process. Thankfully, these were caught in the review process before we submitted the package for the first official release on CRAN.
For example, it is good practice to use an API key when working with APIs, to let the owners track usage. rromeo provides several ways to set up an API key after registering it: a key argument in all functions, setting up an environment variable in the session, using an .Renviron or an .Rprofile file. These 4 options are summarized in the API key vignette. Our first explanation on how to setup an API key was difficult to understand and was located in the function help files. We thank both our reviewers, Philipp Ottolinger and Bruna Wundervald, for encouraging us to write a full vignette regarding API keys. Thanks to their comments we also wrote the rr_auth() function that writes the key as an environment variable. There may be room for improvement regarding the security of the API key but the access to the API is probably not very sensitive in our case.
Similar to setting up an API key, setting a user-agent when doing web-scraping is good practice to let the owners of the website/API from which kind of software the requests come from. Bruna Wundervald pointed out that we had forgotten to setup a user-agent even though it was specified in the httr best practices vignette. We tweaked our requests using httr::add_headers("user-agent" = ...) to add a user-agent with a custom defined string that links to the GitHub page of the package and returns its version number.
To check that the user-agent was well defined we used the awesome website https://httpbin.org/ which is a simple HTTP request & response service. It is very useful to test prototype queries and make sure you get back what you wanted. We used https://httpbin.org/user-agent with our custom specified header to make sure we had set up the user-agent correctly. We do recommend using httpbin.org services to prototype requests and test the passing of arguments.
Because all packages are in the end made for users, it is always important to think about the interface you want to expose to your users. We first had a single function rr_journal_name() with a Boolean argument called multiple. Based on the value of this argument, the result of rr_journal_name() could be wildly different. Both our reviewers noticed it was unexpected from a naive user point of view and suggested to split the function in two different functions that had consistent output: rr_journal_name() would return all the information regarding policies and rr_journal_find() would return simply the title as well as the ISSN of the journal if found. The new outlook of independent reviewers helped us take a step back regarding the functions we were exposing to the users.
 Giving back to the community
Giving back to the community
As we mentioned previously, our main source of attraction towards rOpenSci was its thriving community. We’ve always been convinced that collaboration produced the best scientific output. Because of this, we were glad to notice that our experience with rromeo could have ripple effects and help other projects in the community.
Filing issues when you find a bug is an easy but efficient way to give back to the community. vcr and webmockr are recent rOpenSci packages that have mainly been used by a handful of developers until now, one of them being rOpenSci co-founder Scott Chamberlain. Scott often uses crul in his API packages while we used httr for rromeo. We were able to identify issues with vcr and webmockr when used with httr but they were fixed promptly 123.
We also plan to contact the SHERPA/RoMEO API developers to let them know we developed rromeo and have their feedback on the package.
 The future
The future
rromeo is both on GitHub and CRAN now and seems quite stable. The SHERPA/RoMEO API offers more data regarding paid open access policies (https://www.sherpa.ac.uk/romeo/PaidOA.php) as well as other restrictions on manuscript archival. We are still looking for a suitable format to return this information to users. If you want to get involved, we welcome contributions (look for the issues tagged “help wanted” on GitHub).
We enjoyed the experience of contributing to rOpenSci, benefiting from great recommendations by the editor and reviewers, as well as discovering bugs in lesser used packages along the way.
We are now moving to another package that we plan to submit to rOpenSci. It is also an API package so we’ll be using some of the knowledge we got from developing rromeo. We hope to be back soon on the rOpenSci blog to talk about that ;)
 Acknowledgements
Acknowledgements
We would like to thank both of the reviewers Philipp Ottolinger and Bruna Wundervald as well as the editor Scott Chamberlain for their input to improve the documentation and the package. We also want to mention Scott Chamberlain for building so many in tools to make the interactions with API within R easier (vcr, webmockr, crul, as well as many API packages that can be used as examples for developing your own API package).

